When LLSM is combined with 4X Expansion Microscopy (already in place in NINC), it is the only technique which allows nanoscale imaging of millimeter scale volumes of brain tissue at acquisition rates which are amenable to comparison of genotypes or other groups. LLSM enables super-resolution molecular characterization of imaged tissues. It is the histological complement to the iMAP in vivo techniques. Data outputs include identification and quantification of synaptic partners, spines, spine morphologies, and neuronal tracing and myelination analysis with nanoscale resolution over mm of tissue.
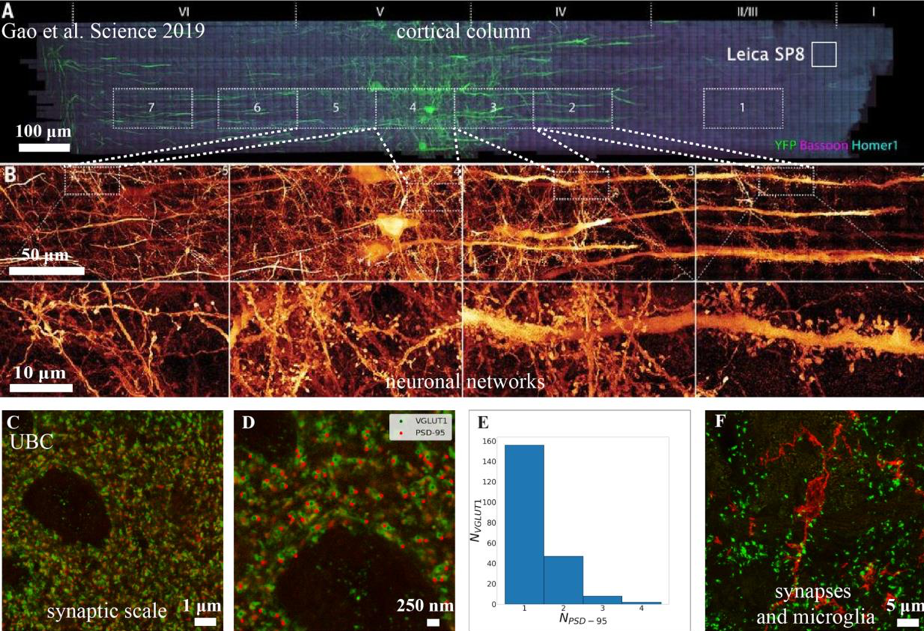
Expansion microscopy (ExM) of mouse cortex: Adapted from Gao et al. (2019). (a) Shows a 1900x280x70μm piece of mouse somatosensory cortex image by lattice light sheet microscopy (LLSM). White box labelled “Leica SP8” indicates the tissue a modern, galvo-based, point scanning confocal would image in the same time as LLSM with favourable confocal imaging conditions (~0.6 million voxels per second for the SP8 versus 100 million voxels per second for the LLSM). (b) Shows the detail throughout the tissue. Results obtained at UBC using ExM showing even more detail: (c) Image of synaptic PSD-95 (red) and VGLUT1 (green). Scale bar 1μm. (d) Annotated image of the upper-left quadrant of (c). Scale bar 250nm (e) Number of PSD-95 clusters associated with each VGLUT1 cluster determined by minimum distance, values reflect multiple-synapse boutons agrees well with ultrastructural measures (Sorra et al. (1993)). (f) Z-stack maximum intensity projection showing VGLUT1 (green) and microglia (red, P2Y12 staining).
Key publications
Chen, B.-C. et al. Lattice light-sheet microscopy: Imaging molecules to embryos at high spatiotemporal resolution. Science (80-. ). 346, (2014).
Liu, T. L. et al. Observing the cell in its native state: Imaging subcellular dynamics in multicellular organisms. Science (80-. ). 360, (2018).
Gao, R. et al. Cortical column and whole-brain imaging with molecular contrast and nanoscale resolution. Science (80-. ). 363, (2019).

